The genome structure of Adineta ricciae: still a mystery
Adineta ricciae is one of the bdelloid rotifer species. It was isolated once many years ago and since then it lives in a lab culture. Earlier work on this species indicated 12 chromosomes and approximate total genome size 250 Mbp. Furthermore the genomes of all bdelloid rotifers are thought to be degenerated tetraploids. In Nowell et al. 2018 four bdelloid rotifers species were sequenced (including A. ricciae) and the degenerated tetraploidy was characterized, although in the supplementary materials this peculiar observation was made - the kmer spectra of the A. ricciae genome contain an extra peak. We further elaborated on why is this peculiar observation peculiar in the supplement “S2 Haplotype structure of Adineta ricciae” of our recent preprint. This report with be only rephrasing the supplementary material.
I worked on this observation with Reuben W. Nowell and Timothy Rhyker Ranallo-Benavidez and discussed it with dozens of researchers everywhere, thank you all, it still bugs me.
Observation
The genome assembly shown four homologous sequences detectable within the assembly. The more closely related were diverged ~5%, while the more diverged (previously called ohnologs) ~25%. With this divergence, we don’t expect any kmers to be shared between the ploidies and therefore we expect a kmer-wise diploid. Hence following model is in a complete agreement with the assembly and all the previous reports
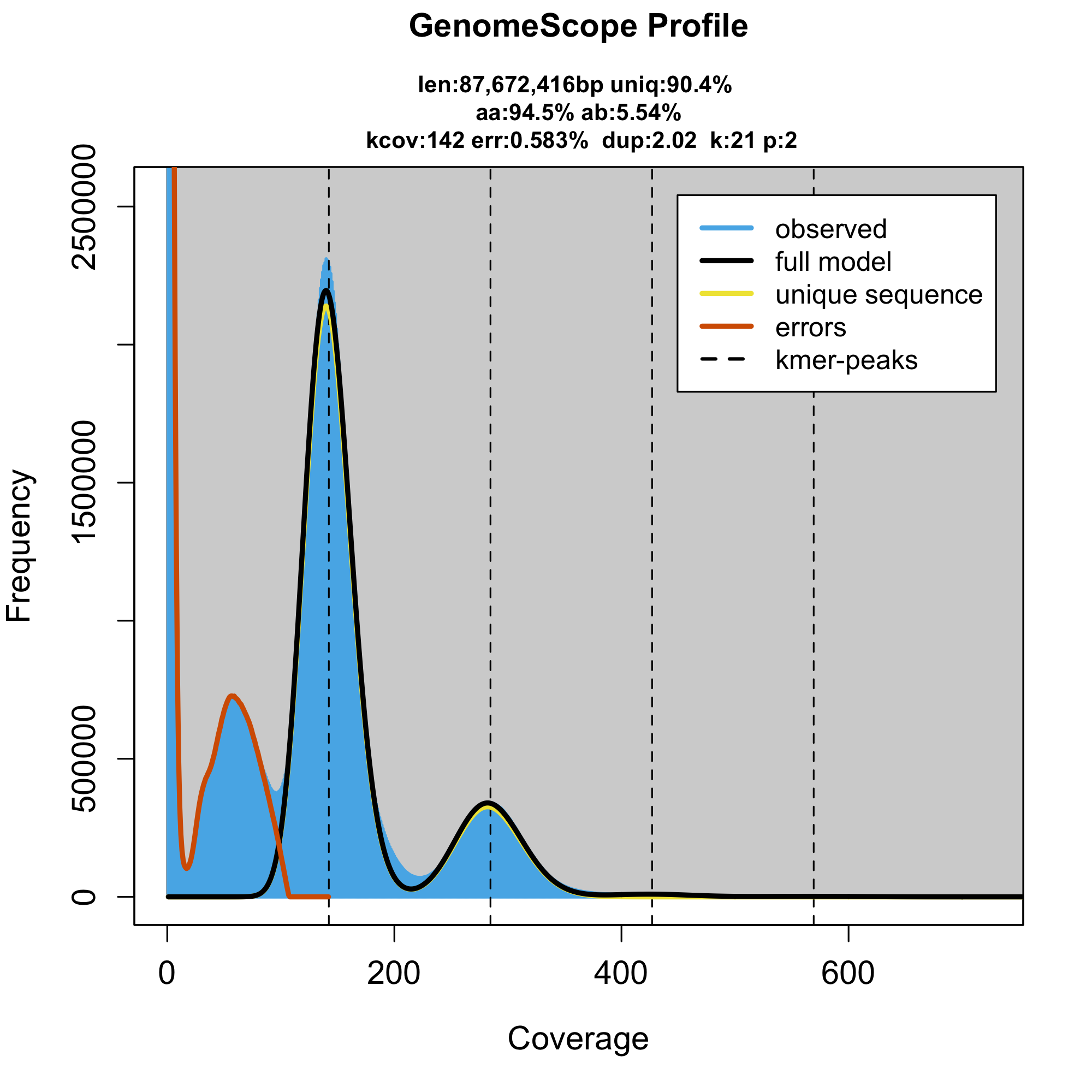
problem is that it ignores the smaller peak around 70x. To figure if the smaller peak contains real genomic kmers we create a smudgeplot, technique that identifies in the kmer set kmers that are similar to each other (one SNP distant), supposedly representing homologous kmers (either heterozygous loci or paralogs). Indeed, the kmers from the 70x peak pair form kmers pairs (rejecting contamination) and further pair with genomic kmers, altogether indicating that the smaller peak must be part of the A. ricciae genome
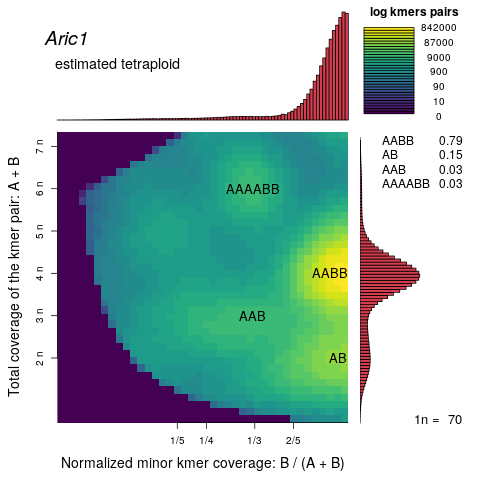
However, the corresponding tetraploid GenomeScope model
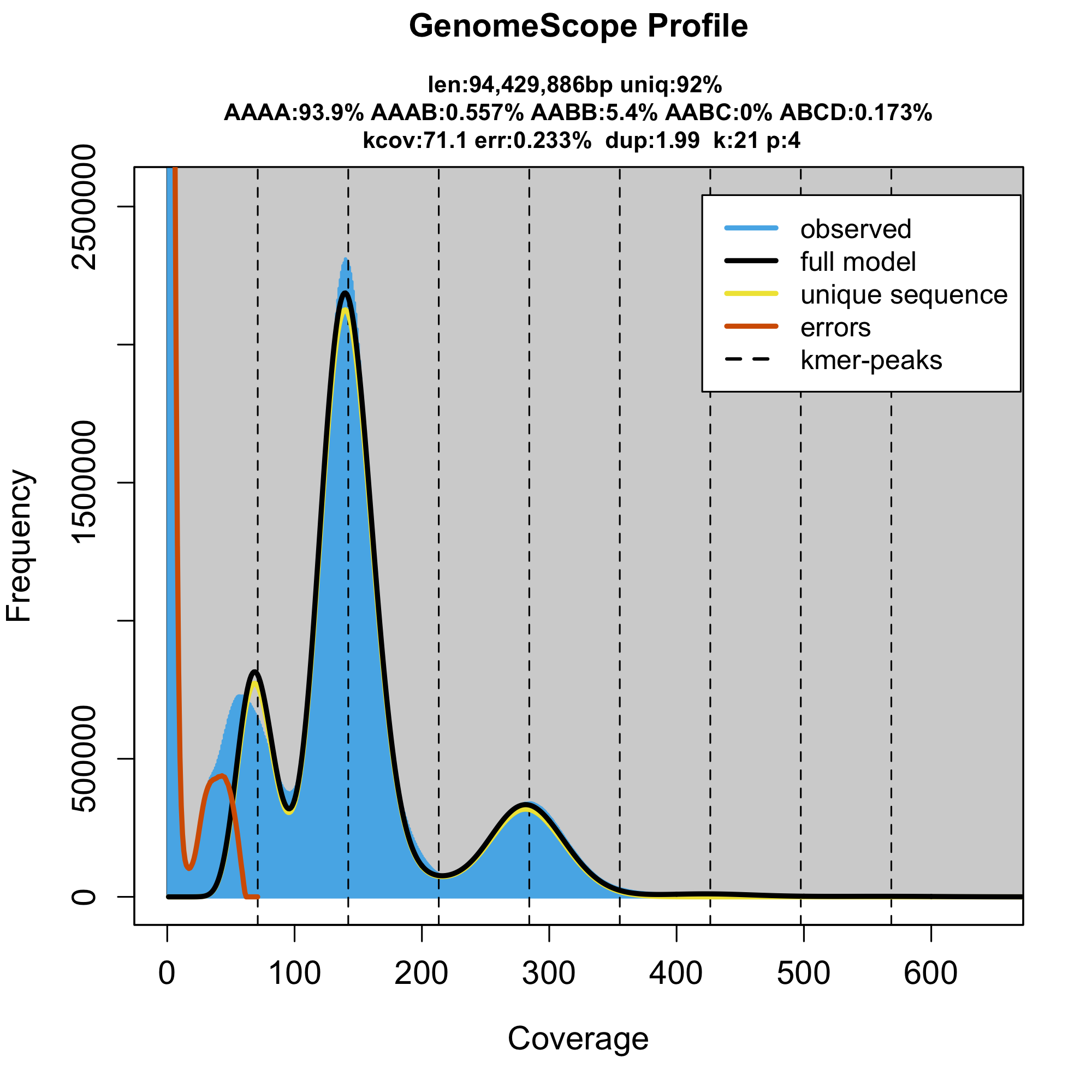
is incompatible with the degenerated tetraploidy. Tetraploid model indicate degenerated octoploidy, that is hardly compatible with the observed 12 chromosomes and furthermore the total DNA content of a cell is supposed to be much smaller than 90 Mbp * 8.
Explanation
There is currently no satisfactory explanation of the observation. Both contradictory sources present solid evidences. Furthermore, it is the same lab culture of the species used for genomics analyses and previous cytology, therefore we can not blame it to population variability either.
As we run out of sane explanations, we might throw some crazier. What if there was a whole genome duplication between the cytology decade ago and the recent genome sequencing? The species is ameiotic, therefore most of the problems associated with WGA are not there.
Alternatively, different cell lines of the rotifers could have different ploidy levels. The lines used for the cytology could have experience some sort of genome exclusion.
Finally, we have seen a third of the honey bee genome missing, perhaps would be reckless to dismiss all the potential technical problems.
Methods
Used data:
- the rotifer sequencing, SRA/EDA id:
ERR2135445
Used software:
- KMC to generate kmer histogram
- GenomeScope for genome profiling
- Smudgeplot to make smudgeplot